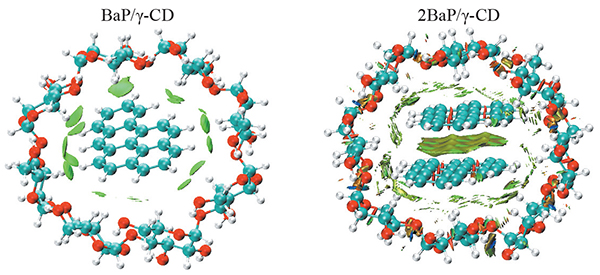
Molecular DFT investigation on the inclusion complexation of benzo[a]pyrene with γ-cyclodextrin
Аннотация
The complexation process between benzo[a]pyrene (BaP) and γ-cyclodextrin (γ-CD) was computationally studied using DFT methodology. Several initial configurations of the interaction of BaP with γ-CD were explored to determine the most stable inclusion complex. According to the calculated complexation energies, the BaP/γ-CD complex is found to be the most favorable energetically when the BaP guest is totally entrapped into γ-CD cavity. The inclusion process involving the encapsulation of two guests BaP in the cavity of γ-CD is also thermodynamically favored. Van der Waals interactions play a determinant role in stabilizing BaP/γ-CD and 2BaP/γ-CD complexes.
Литература
Katsoyiannis A., Breivik K. Environ. Pollut. 2014, 184, 488-494. https://doi.org/10.1016/j.envpol.2013.09.028
Zhou H., Wu C., Onwudili J.A., Meng A., Zhang Y., Williams P.T. Waste Manage. 2015, 36, 136-146. https://doi.org/10.1016/j.wasman.2014.09.014
Rengarajan T., Rajendran P., Nandakumar N., Lokeshkumar B., Rajendran P., Nishigaki I. Asian Pac. J. Trop. Biomed. 2015, 5, 182-189. https://doi.org/10.1016/S2221-1691(15)30003-4
Wang, L., Zhang, S., Wang, L., Zhang, W., Shi, X., Lu, X., Li, X. Int. J. Environ. Res. Public Health. 2018, 15, 607. https://doi.org/10.3390/ijerph15040607
Chang Y., Siddens L.K., Heine L.K., Sampson D.A., Yu Z., Fischer K.A., Löhr C.V., Tilton S.C. Toxico.l Appl. Pharmacol. 2019, 379, 114644. https://doi.org/10.1016/j.taap.2019.114644
Chang Y., Huynh C.T.T., Bastin K.M., Rivera B.N., Siddens L.K., Tilton S.C. Toxicol. In Vitro 2020, 104991. https://doi.org/10.1016/j.tiv.2020.104991
Bolden A.L., Rochester J.R., Schultz K., Kwiatkowski C.F. Reprod. Toxicol. 2017, 73, 61-74. https://doi.org/10.1016/j.reprotox.2017.07.012
Cathey A.L., Watkins D.J., Rosario Z.Y., Vélez Vega C.M., Loch-Caruso R., Alshawabkeh, A.N., Cordero J.F., Meeker J.D. Sci. Total Environ. 2020, 141581. https://doi.org/10.1016/j.scitotenv.2020.141581
Barnes J.L., Zubair M., John K., Poirier M.C., Martin F.L. Biochem. Soc. Trans. 2018, 46,1213-1224. https://doi.org/10.1042/BST20180519
De la Rosa J.M., Sánchez-Martín A.M., Campos P., Miller A.Z. Sci. Total Environ. 2019, 667, 578-585. https://doi.org/10.1016/j.scitotenv.2019.02.421
Cook J.W., Hewett C.L., Hieger I. J. Chem. Soc. 1933, 106, 395. https://doi.org/10.1039/jr9330000395
Phillips D.H. Nature 1983, 303(5917), 468-472. https://doi.org/10.1038/303468a0
Nebert D.W., Shi Z., Galvez-Peralta M., Uno S., Dragin N. Mol. Pharmacol. 2013, 84, 304-313. https://doi.org/10.1124/mol.113.086637
Honda M., Suzuki N. Int. J. Environ. Res. Public Health 2020, 17, 1363. https://doi.org/10.3390/ijerph17041363
IARC. IARC. Monogr. Eval. Carcinog. Risks Hum. 2012, 100F, 111-144.
Nisbet I.C.T., LaGoy P.K. Regul. Toxicol. Pharmacol. 1992, 16, 290-300. https://doi.org/10.1016/0273-2300(92)90009-X
Petry T., Schmid P., Schlatter C. Chemosphere 1996, 32, 639-648. https://doi.org/10.1016/0045-6535(95)00348-7
Ohiozebau E., Tendler B., Codling G., Kelly E., Giesy J.P., Jones P.D. Environ. Geochem. Health. 2016, 39, 139-160. https://doi.org/10.1007/s10653-016-9815-3
Song Y., Nahrgang J., Tollefsen K.E. Sci. Total Environ. 2019, 653, 176-189. https://doi.org/10.1016/j.scitotenv.2018.10.261
Boente C., Baragaño D., Gallego J. R. Environ. Pollut. 2020, 266, 115341. https://doi.org/10.1016/j.envpol.2020.115341
Choong C.E., Wong K.T., Yoon S.Y., Kim, H., Shin, M., Chang Y.Y., Yang J.K., Kim S.H., Jeon B.H., Yoon Y., Jang M. J. Clean. Prod. 2021, 278, 123425. https://doi.org/10.1016/j.jclepro.2020.123425
Morales P., Cáceres M., Scott F., Díaz-Robles L., Aroca G., Vergara-Fernández A. Appl. Microbiol. Biotechnol. 2017, 101, 6765-6777. https://doi.org/10.1007/s00253-017-8400-8
Veignie E., Rafin C., Landy D., Fourmentin S., Surpateanu G. J. Hazard. Mater. 2009, 168, 1296-1301. https://doi.org/10.1016/j.jhazmat.2009.03.012
Greene L., Elzey B., Franklin M., Fakayode S.O. Spectrochim. Acta 2017, 174, 316-325. https://doi.org/10.1016/j.saa.2016.11.047
Szejtli, J. Chem. Rev. 1998, 98, 1743-1753. https://doi.org/10.1021/cr970022c
Tian B., Xiao D., Hei T., Rui Ping R., Shiyao Huac S., Liuc J. Polym Int. 2020, 69, 597-603. https://doi.org/10.1002/pi.5992
Carneiro S., Costa Duarte F., Heimfarth L., Siqueira Quintans J., Quintans-Júnior L., Veiga Júnior V., Neves de Lima Á. Int. J. Mol. Sci. 2019, 20, 642. https://doi.org/10.3390/ijms20030642
Sayede A., Ponchel A., Filardo G., Galia A., Monflier E. J. Mol. Struct. 2006, 777, 99-106. https://doi.org/10.1016/j.theochem.2006.08.033
Bednarek E., Bociana W., Michalska K. J. Pharm. Biomed. Anal. 2019, 169, 170-180. https://doi.org/10.1016/j.jpba.2019.02.031
Westerberg G., Wiklund L. J. Pharm. Sci. 2005, 94, 114-119. https://doi.org/10.1002/jps.20198
Liu T., Ding K., Guo G., Yang F., Wang L. Chem Ecol. 2018, 34, 519-531. https://doi.org/10.1080/02757540.2018.1462343
Yang M., Wang Y., Wang H. Electrochim. Acta 2015, 169, 7-12. https://doi.org/10.1016/j.electacta.2015.04.057
Patonay G., Warner I.M. J. Incl. Phenom. Macrocycl. Chem. 1991, 11, 313-322. https://doi.org/10.1007/BF01041410
Woodberry R., Ransom S., Chen F.M. Anal. Chem. 1988, 60, 2621-2625. https://doi.org/10.1021/ac00174a017
Harata K. Bull. Chem. Soc. Jpn. 1987, 60, 2763-2767. https://doi.org/10.1246/bcsj.60.2763
Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. J. Cheminform. 2012, 4, 17. https://doi.org/10.1186/1758-2946-4-17
Neese F., The ORCA program system, Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2012, 2, 73-78. https://doi.org/10.1002/wcms.81
Neese F., Software update: the ORCA program system, version 4.0. WIREs Comput. Mol. Sci. 2017, 8, e1327. https://doi.org/10.1002/wcms.1327
Liu L., Guo Q.X. J. Incl. Phenom. Macrocycl. Chem. 2004, 50, 95-103. https://doi.org/10.1007/s10847-003-8847-3
Takano Y., Houg K.N. J. Chem. Theory Comput. 2005, 1, 70-77. https://doi.org/10.1021/ct049977a
Becke A.D. J. Chem. Phys. 1997, 107, 8554. https://doi.org/10.1063/1.475007
Grimme S., Ehrlich S., Goerigk L. J. Comput. Chem. 2011, 32, 1456-1465. https://doi.org/10.1002/jcc.21759
Grimme S., Antony J., Ehrlich S., Krieg H. J. Chem. Phys. 2010, 132, 154104. https://doi.org/10.1063/1.3382344
Weigend F., Ahlrichs R. Phys. Chem. Chem. Phys. 2005, 7, 3297. https://doi.org/10.1039/b508541a
Kruse H., Grimme S. J. Chem. Phys. 2012, 136, 154101. https://doi.org/10.1063/1.3700154
Weigend F. Phys. Chem. Chem. Phys. 2006, 8, 1057-1065. https://doi.org/10.1039/b515623h
Eichkorn K., Treutler O., Öhm H., Häser M., Ahlrichs R. Chem. Phys. Lett., 1995, 242, 652-660. https://doi.org/10.1016/0009-2614(95)00838-U
Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/ Accessed date: October 13 2018.
Assaba I.M., Rahali S., Belhocine Y., Allal H. J. Mol. Struct. 2021, 1227, 129696. https://doi.org/10.1016/j.molstruc.2020.129696
Belhocine Y., Bouhadiba A., Rahim M., Nouar L., Djilani I., Khatmi D-E. Macroheterocycles 2018, 11, 203-209. https://doi.org/10.6060/mhc170829b
de Sousa F.B., Leite Denadai A.M., Lula I.S., Nascimento Jr C.S., Fernandes Neto N.S.G., Lima A.C., de Almeida W.B., Sinisterra R.D. J. Am. Chem. Soc. 2008, 130, 8426-8436. https://doi.org/10.1021/ja801080v
Venkatesh G., Sivasankar T., Karthick M. Rajendiran N. J. Incl. Phenom. Macrocycl. Chem. 2013, 77, 309-318. https://doi.org/10.1007/s10847-012-0248-z
Ammouchi N., Allal H., Belhocine Y., Bettaz S., Zouaoui E. J. Mol. Liq. 2020, 300, 133-145. https://doi.org/10.1016/j.molliq.2019.112309
Elistratova M.A., Zakharova I.B., Kvyatkovskii O.E. Macroheterocycles 2019, 12, 370-374. https://doi.org/10.6060/mhc190552e
Fifere A., Marangoci N., Maier S., Coroaba A., Maftei D., Pinteala M. Beilstein J. Org. Chem. 2012, 8, 2191-2201. https://doi.org/10.3762/bjoc.8.247
Nagarajan V., Chandiramouli R. Appl. Surf. Sci. 2015, 344, 65-78. https://doi.org/10.1016/j.apsusc.2015.03.069
Allal H., Belhocine Y., Rahali S., Damous M., Ammouchi N. J. Mol. Model. 2020, 26, 128. https://doi.org/10.1007/s00894-020-04388-3
Contreras-García J., Boto R.A., Izquierdo-Ruiz F., Reva I., Woller T., Alonso M. Theor. Chem. Acc. 2016, 135, 242. https://doi.org/10.1007/s00214-016-1977-7
Saleh G., Gatti C., Presti L.L. Comput. Theor. Chem. 2012, 998, 148-163. https://doi.org/10.1016/j.comptc.2012.07.014
Lefebvre C., Rubez G., Khartabil H., Boisson J.C., Contreras-García J., Hénon E. Phys. Chem. Chem. Phys. 2017, 19, 17928-17936. https://doi.org/10.1039/C7CP02110K
Lu T., Chen F. J. Comput. Chem. 2012, 33, 580-592. https://doi.org/10.1002/jcc.22885
Humphrey W., Dalke A., Schulten K. J. Mol. Graph. 1996, 14, 33-38. https://doi.org/10.1016/0263-7855(96)00018-5